by Kenneth W. Krause.
Kenneth W. Krause is a contributing editor and “Science Watch” columnist for the Skeptical Inquirer. Formerly a contributing editor and books columnist for the Humanist, he writes regularly for Skeptic magazine as well.
Poised to transform the world as we know it, a new gene-editing system has bioethicists wringing their hands, physicians champing at the bit, and researchers dueling with demons.
Is it possible to overstate the potential of a new technology that efficiently and cheaply permits deliberate, specific, and multiple genomic modifications to almost anything biological? What if that technology was also capable of altering untold future generations of nearly any given species—including the one responsible for creating it? And what if it could be used, for better or worse, to rapidly exterminate entire species?
Certain experts have no intention of veiling their enthusiasm, or their unease. Consider, for example, biologist David Baltimore, who recently chaired an international summit dedicated primarily to the technology’s much-disputed ethical implications. “The unthinkable has become conceivable,” he warned his audience in early December. Powerful new gene-editing techniques, he added, have placed us “on the cusp of a new era in human history.”
If so, it might seem somewhat anticlimactic to note that Science magazine has dubbed this technology its “Breakthrough of the Year” for 2015, or that its primary developers are widely considered shoo-ins for a Nobel Prize—in addition, that is, to the US$3 million Breakthrough Prize in Life Sciences already earned by two such researchers. All of which might sound trifling compared to the billions up for grabs following imminent resolution of a now-vicious patent dispute.
Although no gene-editing tool has ever inspired so much drama, the new technology’s promise as a practical remedy for a host of dreadful diseases, including cancer, remains foremost in researchers’ minds. Eager to move beyond in vitro and animal model applications to the clinical setting, geneticists across the globe are quickly developing improved molecular components and methods to increase the technology’s accuracy. In case you haven’t heard, a truly profound scientific insurrection is well underway.
Adapting CRISPR-Cas9.
Think about a film strip. You see a particular segment of the film that you want to replace. And if you had a film splicer, you would go in and literally cut it out and piece it back together—maybe with a new clip. Imagine being able to do that in the genetic code, the code of life.—biochemist Jennifer Doudna (CBS News 2015).
Genetic manipulation is nothing new, of course. Classic gene therapy, for example, typically employs a vector, often a virus, to somewhat haphazardly deliver a healthy allele somewhere in the patient’s genome, hopefully to perform its desired function wherever it settles. Alternatively, RNA interference selects specific messenger RNA molecules for destruction, thus changing the way one’s DNA is transcribed. Interference occurs, however, only so long as the damaging agent remains within the cell.
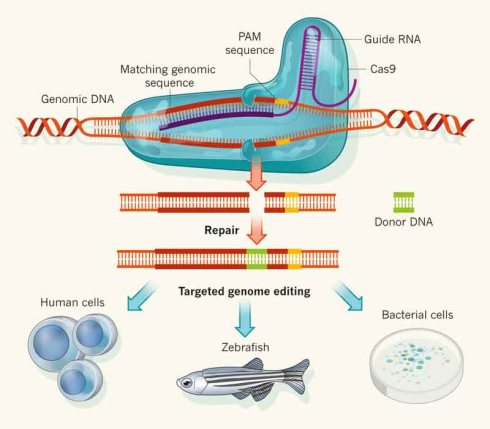
Contemporary editing techniques, on the other hand, allow biologists to actually alter DNA—the “code of life,” as Doudna suggests—and to do so with specific target sequences in mind. The three major techniques have much in common. Each involves an enzyme called a programmable nuclease, for example, which is guided to a particular nucleotide sequence to cleave it.
Then, in each case, the cell’s machinery quickly repairs the double-stranded break in one of two ways. Non-homologous end joining for gene “knock out” results when reconstruction, usually involving small, random nucleotide deletions or insertions, is performed only by the cell. Here, the gene’s function is typically undermined. By contrast, homology-directed repair for gene “knock in” occurs when the cell copies a researcher’s DNA repair template delivered along with the nuclease. In this case, the cleaved gene can be corrected or a new gene or genes can be inserted (Corbyn 2015).
But in other ways, the three editing techniques are very distinct. Developed in the late 1990s and first used in human cells in 2005, zinc-finger nucleases (ZFN) attach cutting domains derived from the prokaryote Flavobacterium okeanokoites to proteins called zinc fingers that can be customized to recognize certain three-base-pair DNA codes. Devised in 2010, transcription activator-like effector nucleases (TALENs) fuse the same cutting domains to different proteins called TAL effectors. For both ZFN and TALENs, two cutting domains are necessary to cleave double-stranded DNA (Maxmen 2015).
The third and most revolutionary editing technique, and subject of this paper, consists of clustered regularly interspaced short palindromic repeats (CRISPR) and a CRISPR-associated protein-9 nuclease (Cas9). Introduced as an exceptionally precise editing technique in 2012 by Doudna at the University of California, Berkeley, and microbiologist Emmanuelle Charpentier at the Max Planck Institute for Infection Biology in Berlin, CRISPR-Cas9 is actually the bacterium Streptococcus pyogenes’ adaptive immune system that confers resistance to foreign elements, like phages and plasmids.
CRISPR thus refers to short bits of DNA seized from invading viruses and stored in the bacterium’s own genome for future reference, and Cas9 is the enzyme S. pyogenes uses to cleave a subsequent invader’s double helix. In other words, in its native setting, CRISPR-Cas9 is the system a certain bacterium uses to recognize and disable common biological threats. Unlike ZFN and TALENs, CRISPR-Cas9 does not rely on the F. okeanoites cutting domain and, as such, can cleave both strands of an interloper’s double helix simultaneously with a single Cas9 enzyme.
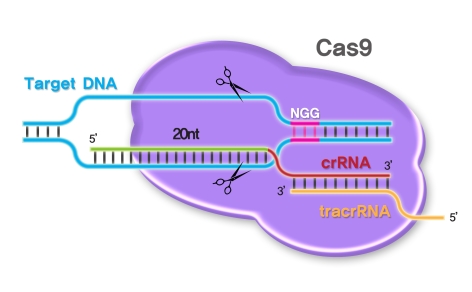
But what makes the CRISPR system so special, in part, and so adaptable to the important task of gene-editing, is its relative simplicity. Only three components are required to achieve site-specific DNA recognition and cleavage. Both a CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA) are needed to guide the Cas9 enzyme to its target sequence. What Doudna and Charpentier revealed six years ago, however, were the seminal facts that an even simpler, two-component system could be developed by combining the crRNA and tracrRNA into a synthetic single guide RNA (sgRNA), and that researchers could readily modify a sgRNA’s code to redirect the Cas9 enzyme to almost any preferred sequence (Jinek et al. 2012). Today, a biologist wanting to edit a specific sequence in an organism’s genome can quickly and cheaply design a sgRNA to match that sequence, order it from a competitive manufacturer for US$65 or less, and have it delivered in the mail (Petherick 2015).
None of which is to suggest that a CRISPR system is always the best tool for the gene-editing job, at least not yet. Critically, CRISPR-Cas9 is relatively easy to program and remains the only technique allowing researchers to “multiplex,” or edit several genomic sites simultaneously. But TALENs have the longest DNA recognition domains and, thus, tend so far to result in the fewest “off-target effects,” which occur when nucleotide sequences identical or similar to the target are cut unintentionally. And ZFNs are much smaller than either TALENs or CRISPR-Cas9, especially the most popular version derived from S. pyogenes, and are therefore more likely to fit into the tight confines of an adeno-associated virus (AAV)—currently the most promising vector for the delivery of gene-editing therapies.
Even so, CRISPR research continues to progress at breakneck speed. In 2014, the number of gene-editing kits ordered from Addgene, a supplier based in Cambridge, Massachusetts, for research using ZFN and TALENs totaled less than 1000 and less than 2000, respectively. During that same year—only two years after the new technology was introduced, the number of kits ordered for CRISPR research totaled almost 20,000 (Corbyn 2015). More importantly, rapidly increasing orders seem to have translated into significant results. As 2015 ended and a new year began, new studies announcing the creation of smaller guide RNAs and, especially, the reduction of off-target effects began to dominate science headlines.
Building a Better Mousetrap.
At some point everyone needs to decide how specific is specific enough. The idea that you would make a tool that has absolutely no off-target effects is a little too utopian.—bioengineer Charles Gersbach (Ledford 2016).
It’s cheap, easy to use, and remarkably efficient, but CRISPR-Cas9 is not perfect. In early experiments, in fact, pathologist Keith Joung at the Massachusetts General Hospital in Boston, discovered that his enzymes were cutting unintended as often as targeted sequences (Servick 2016). The U.S. Food and Drug Administration has yet to announce requirements for clinical use of the new technology. But to help future clinicians safely repair defective, disease-causing genes, for example, researchers are exploring various means of reducing off-target effects that could harm patients in any number of ways, including through uncontrolled cellular growth and cancer.
A CRISPR-Cas9 system “licenses” a DNA sequence for cleavage through a two-stage recognition process (Bolukbasi et al. 2016). Even the most basic details are somewhat technical, of course, but very illuminating. First, a Cas9-sgRNA complex will attach and remain attached to a DNA sequence only if an appropriate protospacer-adjacent motif (PAM) is nearby. PAM sequences are very short, often only a few base-pairs long. In the case of an S. pyogenes Cas9, an NGG PAM is much-preferred, but NAG and NGA PAMs are sometimes inefficiently recognized (“N” represents any nucleobase followed by two guanine, or “G” nucleobases).
Second, and only if an appropriate PAM is recognized, the sgRNA will interrogate the neighboring DNA sequence through Watson-Crick base pairing in a 3′-to-5′ direction. For an S. pyogenes Cas9, the guide sequence will measure twenty nucleotides long. If the 3′ end of the programmed guide sequence is complementary to the DNA sequence near the PAM element, “R-loop” formation is initiated. In zipper-like fashion, further complementarity of the DNA is assessed through extension of the R-loop. If a complete target sequence is confirmed, allosteric activation of the Cas9 enzyme—actually, activation of Cas9’s two nuclease domains, RuvC and HNH—will result in dual cleavage and, accordingly, a complete double-stranded break in the target sequence.
Unsurprisingly, then, the specificity of a CRISPR-Cas9 system is determined in two ways. In large part, off-target effects are managed through careful design of the sgRNA. Ideally, the guide sequence would match the target sequence perfectly, and show no homology elsewhere in the genome. More realistically, however, at least partial homology will often occur at other genomic sites where, unfortunately, off-target cleavage could ensue. Researchers have developed algorithms that help predict sufficient homology, but have yet to clearly and comprehensively define how closely guide and DNA sequences must harmonize before licensing occurs. Nevertheless, nuclease activity has been observed at off-target sites displaying up to four or five nucleotide mismatches.
So, careful design of the sgRNA is critical. But one team of researchers, including Joung, recently confirmed that truncating the guide sequence can also help (Fu et al. 2014). Shortening their guides to as few as seventeen nucleotides, instead of the usual twenty, Joung’s group was able to not only decrease nuclease activity at many off-target sites, but to preserve nearly thorough activity at the majority of intended sites as well.
Other groups have achieved similar success by inactivating one of the two nuclease domains, thus creating a “nickase” that cleaves only one strand of the target sequence (Ran et al. 2013). Here, a double-stranded break can still be achieved by joining two Cas9 nickases with two different sgRNAs targeting adjacent sites on opposing DNA strands. Importantly, the obligatory use of two active nickases decreases the likelihood of off-target cleavage.
Perhaps the latest and most significant progress in this area, however, has been achieved through modification of the unaltered, or “wild-type,” Cas9 nuclease. Last December, for example, synthetic biologist Feng Zhang at the Broad Institute of MIT and Harvard University announced that he and his colleagues had engineered the Cas9 to render it less likely to act at genomic sites presenting mismatches between RNA guides and DNA targets (Slaymaker et al. 2015). Appropriately, Zhang dubbed his new enzyme an “enhanced specificity” S. pyogenes Cas9, or eSpCas9 for short.
Knowing that negatively charged DNA binds to a positively charged groove in the Cas9 enzyme, Zhang’s team predicted that by replacing only a few among the 1400 or so positively charged amino acids with neutral equivalents they could temper the wild-type Cas9’s enthusiasm for binding to and cutting off-target sites. They created and tested several new versions of enzyme that reportedly reduced unintended activity at least tenfold, while maintaining robust on-target cleavage.
Earlier this year, however, Joung and colleagues claimed to have bested Zhang’s results by bringing “off-target-effects to levels where we can no longer detect them, even with the most sensitive methods” (McGreevey 2016). Like Zhang, Joung focused on points of interaction between Cas9 and DNA sequences. His team created fifteen new enzyme variants by replacing up to four long amino acid side-chains that bind to DNA with shorter chains that do not (Kleinstiver et al. 2016).
Joung then tested each of his Cas9 variants in human cells, and found that one three-substitution and one four-substitution version rejected mismatched sites while maintaining full on-target activity. The latter variant, subsequently named SpCas9-HF1—“HF” denoting “high-fidelity,” induced targeted activity as reliably as a wild-type Cas9 when deployed with eighty-five percent of the thirty-seven different guide RNAs tested. Similarly, SpCas9-HF1 generated no detectable off-target mutations with six of seven guide RNAs (and only one mutation with the seventh) compared to twenty-five such effects produced by the wild-type Cas9.
Joung’s group also tested their hi-fi creations at less typical genomic locations that are particularly difficult to control for off-target effects due to the inclusion of repeat sequences. But even there, his supplemental variants, since designated HF2, HF3, and HF4, appeared to eliminate off-target activity that tended to persist following use of the HF1 version.
It’s too early to judge which of these innovations will prove most valuable or, in fact, whether all of them will soon be superseded by modifications or entirely different systems yet to be introduced. But much progress has already been made and, importantly, at this point, many of the foregoing strategies and designs can be used in concert to bring us closer yet to the day when CRISPR gene-editing becomes a clinical convention.
Breaking Barriers.
This is now the most powerful system we have in biology. Any biological process we care about now, we can get the comprehensive set of genes that underlie that process. That was just not possible before.—biochemist David Sabatini (Yong 2015).
CRISPR-Cas9, of course, is only one among many prokaryotic CRISPR systems that could, at some point, prove useful for any number of human purposes. Use of Cas9 variations, however, has already resulted in successes far too numerous to review liberally here. Even so, two recent applications in particular reveal the extraordinary, yet strikingly simple, means by which researchers have achieved previously unattainable outcomes.
In the first, three different teams confronted Duchenne muscular dystrophy (DMD), a terrifying disease that affects about one in every 3500 boys in the U.S. alone (Long et al. 2015, Nelson et al. 2015, and Tabebordbar et al. 2015). DMD typically stems from defects in a gene containing seventy-nine protein-coding exons. If even a single exon suffers a debilitating mutation, the gene can be rendered incapable of producing dystrophin, a vital protein that protects muscle fibers. Absent sufficient dystrophin, both skeletal and heart muscle will deteriorate. Patients usually end up confined to wheelchairs and dead before the age of thirty.
Traditional gene therapy, stem cell treatments, and drugs have proven mostly ineffective against DMD. Scientists have corrected diseased cells in vitro, or in a single organ—the liver. But treating muscle cells throughout the body, including the heart, is a far more daunting task, because they can’t all be removed, treated in isolation, and then replaced. And given current ethical concerns, most researchers are prohibited from even considering the possibility of editing human embryos for clinical purposes.
As such, researchers here decided to employ CRISPR-Cas9 technology to excise faulty dystrophin gene exons in both adult and neonatal mice by delivering it directly into their muscles and bloodstreams using non-pathogenic adeno-associated viruses. AAVs, however, are too small to accommodate the relatively large S. pyogenes Cas9, so each team opted instead to deploy a more petite Cas9 enzyme found in Staphylococcus aureus.
Neither group’s interventions resulted in complete cures. But dystrophin production and muscle strength was restored, and little evidence of off-target effects was observed, in treated mice. One lead researcher later suggested that, although clinical trials could be years away, up to eighty percent of human DMD victims could benefit from defective exon removal (Kaiser 2015).
Remarkably, each of the three teams obtained results comparable to those of the others. Perhaps most impressively, however, these experiments marked the very first instances of using CRISPR to successfully treat genetic disorders in fully-developed living mammals.
But an ever-growing population needs to protect its agricultural products too. Plant DNA viruses, for example, can cause devastating crop damage and economic crises worldwide, but especially in underdeveloped regions including sub-Saharan Africa. More specifically, the tomato yellow leaf curl virus (tomato virus) is known to ravage a variety of tomato breeds, causing stunted growth, abnormal leaf development, and fruit death.
Like DMD, the tomato virus has proven an especially intractable problem. Despite previous efforts to control it through breeding, insecticides targeting the vector, and other engineering techniques, we currently know of no effective means of managing the virus. Undeterred, another group of biologists decided to give CRISPR-Cas9-mediated viral interference a try (Ali et al. 2015).
In this study, the investigators chose to manipulate a species of tobacco plant, well-understood as a model organism, which is similarly vulnerable to tomato virus infection. The experiment was completed in two fairly predictable stages. First, the group designed sgRNAs to target certain tomato virus coding and non-coding sequences and inserted them into different, harmless viruses of the tobacco rattle variety. Second, they delivered the newly loaded rattle viruses into their tobacco plants. After seven days, the plants were exposed to the tomato virus and, after ten more days, they were analyzed for symptoms of infection.
The group agreed that the CRISPR-Cas9 system had reliably cleaved and introduced mutations to the tomato viruses’ genomes. Fortuitously, every plant expressing the system had either abolished or significantly attenuated all symptoms of infection. The investigators concluded further that the technique was capable of simultaneously targeting multiple DNA viruses with a lone sgRNA, and that other transformable plant species, including tomatoes, of course, would be similarly affected.
One can only guess, at this point, how certain interests might receive these and other types of genome-edited crops. Will nations eventually classify them as GMO or, alternatively, as organisms capable of developing in nature? Will applicable regulations focus on the processes or products of modification? Regardless, one can hardly ignore these commodities’ potential windfalls, especially for those in dire need.
Given recent innovations in specificity, for example, CRISPR-based disease research will likely continue to advance quickly toward clinical and other more practical applications. So long as it affects only non-reproductive somatic cells, such interventions should remain largely uncontroversial. Human gametes and embryos, on the other hand, have once again inspired abundant debate and bitter division among experts.
Moralizing Over Science.
Genome editing in human embryos using current technologies could have unpredictable effects on future generations. This makes it dangerous and ethically unacceptable.—Edward Lanphier et al. (2015).
To intentionally refrain from engaging in life-saving research is to be morally responsible for the foreseeable, avoidable deaths of those who could have benefitted.—bioethicist Julian Savulescu et al. (2015).
The results of the first and, so far, last attempt to edit human embryos using CRISPR-Cas9 was published by a team of Chinese scientists on April 18 of last year (Liang et al. 2015). Led by Junjiu Huang, the group chose to experiment on donated tripronuclear zygotes—non-viable early embryos containing one egg and two sperm nuclei—neither intended nor suitable for clinical use. Their goal was to successfully edit endogenous β-globin genes that, when mutated, can cause a fatal blood disorder known as β-thalassemia.
By his own admission, Huang’s outcomes were less than spectacular. Eighty-six embryos were injected with the Cas9 system and a molecular template designed to affect the insertion of new DNA. Of the seventy-one that survived, fifty-four embryos were tested. A mere twenty-eight were successfully spliced and, of those, only four exhibited the desired additions. Rates of off-target mutations were much higher than expected too, and the group would likely have discovered additional unintended cuts had they examined more than the protein-coding exome, which represents less than two percent of the entire human genome.
In all fairness, however, the embryos’ abnormality might have been responsible for much of the total off-target effect. And, of course, Huang was unable to take advantage of many specificity-enhancing upgrades to the CRISPR system yet to be designed at the time of his investigations. In any case, his team acknowledged that their results “highlight the pressing need to further improve the fidelity and specificity” of the new technology, which in their opinions remained immature and unready for clinical applications.
Nevertheless, the Chinese experiment ignited a brawl among both scientists and bioethicists over the prospect of human germline modification with the most powerful and accessible editing machinery ever conceived. Similar quarrels had accompanied the proliferation of technologies involving recombinant DNA, in vitro fertilization, gene therapy, and stem cells, for example. But never had the need to address our capacity to reroute the evolution of societies—indeed, of the entire species—seemed so real and immediate.
Leading experts, including Baltimore and Doudna, had previously met in Napa, California, on January 24, 2015 to discuss the bioethical implications of rapidly emerging technologies. In the end, they “strongly discouraged … any attempts at germline genome modification for clinical application in humans,” urged informed discussion and transparent research, and called for a prompt global summit to recommend international policies (Baltimore et al. 2015). A surge of impassioned literature ensued.
A small group led by Sangamo BioSciences president, Edward Lanphier, was one of the first to weigh in (Lanphier et al. 2015). Calling for a “voluntary moratorium” on all human germline research, Lanphier first expressed concerns over potential off-target effects and the genetic mosaicism that could result, for instance, if a fertilized egg began dividing before all intended corrections had occurred. He also found it difficult to “imagine a situation in which use of human embryos would offer therapeutic benefits over existing and developing methods,” suggesting as well that pre-implantation genetic diagnosis (PGD) and in vitro fertilization (IVF) were far better options than CRISPR for parents carrying the same mutation for a genetic disease. In any case, he continued, with so many unanswered questions, clinicians remained unable to obtain truly risk-informed consent from either parents looking to modify their germlines or from affected future generations. Finally, Lanphier implied that even the best intentions could eventually lead societies down a “slippery slope” toward non-therapeutic genetic enhancement and so-called “designer babies.”
Francis Collins, evangelical Christian and director of the National Institutes of Health (which currently refuses to fund human germline research), expressed similar views regarding the sufficiency of PGD and IVF, the impossibility of informed consent, and non-therapeutic enhancement (Skerrett 2015). Additionally, Collins worries that access to the technology would be denied to the economically disadvantaged and that parents might begin to conceive of their children “more like commodities than precious gifts.” For the director, given the “paucity of compelling cases” in favor of such research, and the significance of the ethical counterarguments, “the balance of the debate leans overwhelmingly against human germline engineering.”
On the other hand, Harvard Medical School geneticist, George Church, urges us to ignore pleas for artificially imposed bans, “encourage the innovators,” and focus more on what he deems the obvious benefits of germline research (Church 2015). Responding to Lanphier and Collins, he argues as well that, without obtaining consent, parents have long exposed future generations to mutagenic forces—through chemotherapy, residence in high-altitudes, and alcohol intake, for example. We have also consistently chosen to enhance our offspring and future generations through mate choice, among many other things. Church also points out that PGD during the IVF procedure is incapable of offering solutions to individuals possessing two copies of a detrimental, dominant allele, or to prospective parents who both carry two copies of a harmful, recessive allele. Moreover, in most instances, PGD cannot be used to avoid more complex polygenic diseases, including schizophrenia. Nor can we presume that new technology costs will always create treatment or enhancement inequities. In fact, according to Church, the price of DNA sequencing, for example, has already plummeted more than three million fold. Finally, germline editing is probably not irreversible, Church contends, and certainly not as error-prone at this point as many have suggested. “Senseless” bans, he concludes, would only “put a damper on the best medical research and instead drive the practice underground to black markets and uncontrolled medical tourism.”
Taking a slightly different tack, Harvard cognitive scientist, Steven Pinker, censures bioethicists generally for getting bogged down in “red-tape, moratoria, or threats of prosecution based on nebulous but sweeping principles such as ‘dignity,’ ‘sacredness,’ or ‘social justice’” (Pinker 2015a). Imploring the bioethical community to “get out of the way” of CRISPR, Pinker reminds them that, once decried as morally unacceptable, vaccinations, transfusions, artificial insemination, organ transplants, and IVF have all proven “unexceptional boons to human well-being.” Further, the specific harms of which moratorium proponents warn, including cancer, mutations, and birth defects, “are already ruled out by a plethora of existing regulations and norms” (Pinker 2015b). In the end, he advises, both scientists and everyday people need and deserve a well-diversified research portfolio. “If you ban something, the probability that people will benefit is zero. If you don’t ban it, the probability is greater than zero.”
Such were among the arguments considered by a committee of twelve biologists, physicians, and ethicists during the December, 2015 International Summit on Human Genome Editing, organized by the U.S. National Academies of Science and Medicine, the Royal Society in London, and the Chinese Academy of Sciences. The Summit was chaired by David Baltimore. Doudna and Charpentier, winners of the US$3 million Breakthrough Prize in Life Sciences, attended with Zhang—a now much-celebrated trio considered front runners for a Nobel Prize, though also entangled through their institutions in a CRISPR patent dispute potentially worth billions of dollars.
After three days of discussion, the Summit’s organizing committee issued a general statement rejecting calls for a comprehensive moratorium on germline research (NAS 2015). The members did, however, advise without exception against the use of edited embryos to establish pregnancy. “It would be irresponsible to proceed,” they added, “with any clinical use of germline editing” until safety and efficacy issues are resolved and there exists “a broad societal consensus about the appropriateness of the proposed application.” In conclusion, the committee called for an “ongoing forum” to harmonize the current global patchwork of relevant regulations and guidelines and to “discourage unacceptable activities.” This forum, the members judged, should consist not only of experts and policymakers, but of “faith leaders,” “public interest advocates,” and “members of the general public” as well.
Wasting little time, the UK’s Human Fertilization and Embryology Authority approved on February 1, 2016, the first attempt to edit healthy human embryos with the CRISPR-Cas9 system. The application was filed last September by developmental biologist, Kathy Niakan, of the Francis Crick Institute in London. Niakan intends to use CRISPR to knock out one of four different genes in a total of 120 day-old, IVF-donated embryos to investigate the roles such genes play in early development.
Her research could help identify genes crucial to early human growth and cell differentiation and, thus, lead to more productive IVF cultures and more informed selection practices. It could also reveal mutations that lead to miscarriages and, one day, allow parents to correct these problems through gene therapy. Following careful observation, Niakan intends to destroy her embryos by the time they reach the blastocyst stage on the seventh day. Under British law, experimental embryos cannot be used to establish pregnancy.
But the human germline is not the only, or even most pressing, subject of CRISPR controversy. Some, for example, warn of the creation of dangerous pathogens and biological warfare (Greely 2016). But many others, including Doudna, urge that we quickly address “other potentially harmful applications … in non-human systems, such as the alteration of insect DNA to ‘drive’ certain genes into a population” (Doudna 2015).
Driving DNA.
Clearly, the technology described here is not to be used lightly. Given the suffering caused by some species, neither is it obviously one to be ignored.—evolutionary geneticist Austin Burt (2003).
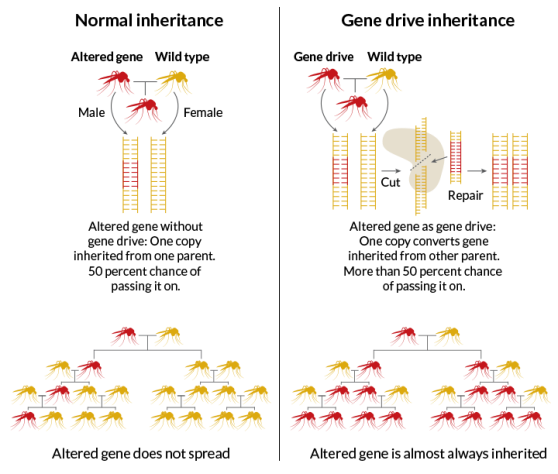
In broad terms, a “gene drive” can be characterized as a targeted contagion intended to spread through a population with exceptional haste. Burt pioneered the technology through his study of transposable elements—“selfish” and often parasitic DNA sequences that exist merely to propagate themselves. Importantly, transposons can circumvent the normal Mendelian rules of inheritance dictating that any given gene has a fifty percent chance of being passed from parent to offspring.
Thirteen years ago, Burt envisioned the use of a microbial transposon-like element called a “homing endonuclease” for humanity’s benefit. When inserted into one chromosome, the endonuclease would cut the matching chromosome inherited from the other parent. The cell would then quickly repair the cut, often using the first chromosome as a template. As such, the assailed sequence in the second chromosome would be converted to the sequence of the selfish element. In a newly fertilized egg, the endonuclease would likewise convert the other parent’s DNA and, eventually, drive itself into the genomes of nearly one-hundred percent of the population.
Burt believes we can use gene drives to weaken or even eradicate mosquito transmitted diseases like malaria and dengue fever. If scientists engineered just one percent of a mosquito population to carry such a drive, he calculates, about ninety-nine percent would possess it in only twenty generations. In fact, Burt announced five years ago that he had created a homing endonuclease capable of locating and cutting a mosquito gene (Windbichler 2011). But his elements were difficult to program for precise application.
Enter CRISPR-Cas9. As we’ve seen, Cas9 is an eager endonuclease and guide RNAs are easy to program and can be quickly synthesized. In April of last year, biologists Valentonio Gantz and Ethan Bier revealed that they had used CRISPR-Cas9 to drive color variation into Drosophila fruit flies (Gantz and Bier 2015). Though they labeled it a “mutagenic chain reaction” at the time, it was the first gene drive ever deployed in a multicellular organism.
Today, researchers sort potential gene drives into two major groups. Replacement drives seek only to displace natural with modified populations. Suppression drives, by contrast, attempt to reduce or even eradicate populations. At this point, no drives have been released into the wild. Nevertheless, researchers have lately designed one of each type to affect mosquitos carrying the deadly human malaria parasite, Plasmodium falciparum.
The first study was led by microbiologist Anthony James, who collaborated on the project with Gantz and Bier (James et al. 2015). Focusing on the prevention of disease transmission, this group engineered Anopheles stephensi mosquitos, highly active in urban India, to carry two transgenes producing antibodies against the malaria parasite, a CRISPR-Cas9-mediated gene drive, and a marker gene. Because the very lengthy payload rendered insertion a challenging process, James was able to isolate only two drive-bearing males among 25,000 larvae. But when mated with wild-type females, these and subsequent transgenic males spread their anti-malaria genes at an impressive rate of 99.5 percent. Transgenic females, on the other hand, processed the drive quite differently and passed it on at near-normal Mendelian ratios.
Despite its overall success, James doesn’t imagine that his team’s replacement drive could eliminate the malaria parasite independently. Instead, he envisions its use to reduce the risk of infection and to compliment other strategies already being employed. Even so, because such drives would not exterminate P. falciparum or its mosquito vector, they would potentially allow the parasite to one day evolve resistance to their transgene components.
The second study’s goal was quite different. Here, molecular biologist, Tony Nolan, along with Burt and others, first identified three genes in the Anopheles gambiae mosquito, active in sub-Saharan Africa, that when mutated cause recessive infertility in females (Hammond et al. 2016). Second, they designed a CRISPR-Cas9 gene drive to target and edit each gene. Following insertion, they bred their transgenic mosquitos with wild-types and found that nearly all female offspring were born infertile. In a subsequent experiment, Nolan released 600 vectors—half transgenic, half wild-type—into a cage. After only four generations, seventy-five percent of the population carried the mutations, exactly what one would expect from an effective gene drive.
A suppression drive like Hammond’s could, in theory, eliminate a parasite’s primary vector. In such a scenario, the parasite might find another means of conveying the disease to humans—more than 800 species of mosquito inhabit Africa alone, for example. But it might not. The loss would also substantially alter the relevant ecosystem. But despite other methods of controlling the disease, malaria still claims more than a half million lives every year, mostly among children under five.
Even in theory, no gene drive is a panacea. They function only in sexually reproducing species, and best in species that reproduce very rapidly. Nor would their effects be permanent—most transgenes would prove especially vulnerable to evolutionary deselection, for example. But neither would they turn out as problematic as some might imagine. They can be easily detected through genome sequencing, for instance, and are unlikely to spread accidentally into domesticated species. And if scientists sought for whatever reason to reverse the effects of a previously released drive, they could probably do so with the release of a subsequent drive.
As Church and others have recently suggested, it “doesn’t really make sense to ask whether we should use gene drives. Rather, we’ll need to ask whether it’s a good idea to consider driving this particular change through this particular population” (Esvelt et al. 2014).
References:
Ali, Z., A. Abulfaraj, Ali Idris, et al. 2015. CRISPR/Cas9-mediated viral interference in plants. Genome Biology 16:238 DOI:10.1186/s13059-015-0799-6.
Baltimore, D., P. Berg, M. Botcham, et al. 2015. A prudent path forward for genomic engineering and germline gene modification. Science 348(6230):36-38.
Bolukbasi, M.F., A. Gupta, and S.A. Wolf. 2016. Creating and evaluating accurate CRISPR-Cas9 scalpels for genomic surgery. Nature Methods 13(1):41-50.
Burt, A. 2003. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proceedings of the Royal Society B 270:921-928.
CBS News. 2015. Could Revolutionary Gene-editing Technology End Cancer? Available online at http://www.cbsnews.com/news/crispr-jennifer-doudna-gene-editing-technology-diseases-dangers-ethics/; accessed January 25, 2016.
Church, G., 2015. Encourage the innovators. Nature 528:S7.
Corbyn, Z. 2015. Biology’s big hit. Nature 528:S4-S5.
Doudna, J. 2015. Embryo editing needs scrutiny. Nature 528:S6.
Esvelt, K., G. Church, and J. Lunshof. 2014. “Gene Drives” and CRISPR Could Revolutionize Ecosystem Management. Available online at http://blogs.scientificamerican.com/guest-blog/gene-drives-and-crispr-could-revolutionize-ecosystem-management/; accessed February 6, 2016.
Fu, Y., J.D. Sander, D. Reyon, et al. 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature Biotechnology 32:279-284.
Gantz, V.M., and E. Bier. 2015a. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science 348(6233):442-444.
Gantz, V.M., N. Jasinskiene, O. Tatarenkova, et al. 2015. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proceedings of the National Academy of Sciences DOI: 10.1073/pnas.1521077112.
Greely, H.T. 2016. Are We Ready for Genetically Modified Animals? Available online at http://www.weforum.org/agenda/2016/01/are-we-ready-for-genetically-modified-animals; accessed February 3, 2016.
Hammond, A. R. Galizi, K. Kyrou, et al. 2016. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nature Biotechnology DOI: 10.1038/nbt.3439.
Jinek, M., K. Chylinski, I. Fonfara, et al. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816-821.
Kaiser, J. 2015. CRISPR Helps Heal Mice With Muscular Dystrophy. Available online at http://www.sciencemag.org/news/2015/12/crispr-helps-heal-mice-muscular-dystrophy; accessed January 30, 2015.
Kleinstiver, B.P., V. Pattanayak, M.S. Prew, et al. 2016. High-fidelity CRISPR-Cas9 nuclease with no detectable genome-wide off-target effects. Nature 529:490-495.
Lanphier, E., F. Urnov, S.E. Ehlen, et al. 2015. Don’t edit the human germline. Nature 519:410-411.
Ledford, H. 2016. Enzyme Tweak Boosts Precision of CRISPR Genome Edits. Available online at http://www.nature.com/news/enzyme-tweak-boosts-precision-of-crispr-genome-edits-1.19114; accessed January 28, 2016.
Liang, P., Y. Xu, X. Zhang, et al. 2015. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell 6(5):363-372.
Long, C., L. Amoasii, A.A. Mireault, et al. 2015. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science DOI: 10.1126/science.aad5725.
Maxmen, A. 2015. Three technologies that changed genetics. Nature 528:S2-S3.
McGreevey, S. 2016. High-fidelity CRISPR. Available online at https://hms.harvard.edu/news/high-fidelity-crispr; accessed January 29, 2016.
National Academies of Science. 2015. International Summit Statement. Available at http://www8.nationalacademies.org/onpinews/newsitem.aspx?RecordID=12032015a; accessed January 2, 2016.
Nelson, C.E., C.H. Hakim, D.G. Ousterout, et al. 2015. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science DOI: 10.1126/science.aad5143.
Petherick, A. 2015. Nature outlook genome editing. Nature 528:S1.
Pinker, S. 2015a. The Moral Imperative for Bioethics. Available online at https://www.bostonglobe.com/opinion/2015/07/31/the-moral-imperative-for-bioethics/JmEkoyzlTAu9oQV76JrK9N/story.html; accessed February 2, 2016.
Pinker, S. 2015b. Steven Pinker Interview. Available online at https://www.ipscell.com/2015/08/stevenpinker/; accessed February 2, 2016.
Ran, F.A., P.D. Hsu, C. Lin, et al. 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154:1380-1389.
Savulescu, J., J. Pugh, T. Douglas, et al. 2015. The moral imperative to continue gene editing research on human embryos. Protein Cell 6(7):476-479.
Servick, K. 2016. Researchers Rein In Slice-happy Gene Editor, CRISPR. Available online at http://www.sciencemag.org/news/2016/01/researchers-rein-slice-happy-gene-editor-crispr; accessed January 28, 2016.
Sherkow, J.S. 2015. The CRISPR Patent Interference Showdown Is On. Available online at https://law.stanford.edu/2015/12/29/the-crispr-patent-interference-showdown-is-on-how-did-we-get-here-and-what-comes-next/; accessed January 29, 2016.
Skerrett, P., 2015. First Opinion. A Debate: Should We Edit the Human Genome? Available online at http://www.statnews.com/2015/11/30/gene-editing-crispr-germline/; accessed February 2, 2016.
Slaymaker, I. M., L. Gao, B. Zetsche, et al. 2015. Rationally engineered Cas9 nucleases with improved specificity. Science 351(6268):84-88.
Tabebordbar, M., K. Zhu, J.K.W. Cheng, et al. 2015. In vivo gene editing in dystrophic mouse and muscle stem cells. Science DOI: 10.1126/science.aad5177.
Windbichler, N., M. Menichelli, P.A. Papathanos, et al. 2011. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature 473:212-215.
Yong, E. 2015. The New Gene-editing Technique that Reveals Cancer’s Weaknesses. Available online at http://www.theatlantic.com/science/archive/2015/11/a-revolutionary-gene-editing-technique-reveals-cancers-weaknesses/417495/; accessed on January 30, 2016.









Un-friggin’-real. Can’t wait to see this stuff happen!
LikeLike
Careful what you ask for. Sounds good here, people can be very irresponsible with power.
LikeLike
Please, no hypothetical doomsday scenarios! Nobody wants to do harm with this technology and evidently everyone wants to do good. The science is well under control and everything is being done above-board. Support a better future, for future’s sake!
LikeLike
Are you really saying that nothing bad could come out of this? Even accidentally? That’s just silly.
LikeLike
What I’m saying, Matt, is that speculation about possible harms is unhelpful. If you want to be a part of the process, do something to help us understand the relevant facts.
LikeLike